PSI-Coffee combines homology extension and consistency based progressive alignment. It is designed for aligning distantly related proteins for which no structural information is available. The requirements of the homology extension (BLAST against nr) make it slower than T-Coffee and Fast M-Coffee. By default, PSI-Coffee uses the EBI web-services and runs remotely (at the EBI) the BLASTs required for the homology extension procedure. PSI-Coffee run via the following command: t_coffee -seq sh3.fasta -mode psicoffee The psicoffee mode produces by five different types of output files:
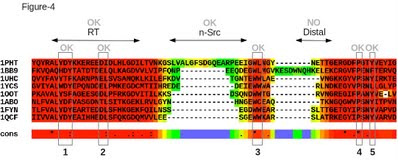
The alignment presented Figure 4 illustrate the advantage of using homology information as PSI-Coffee can accurately align two difficult parts: the n-Src loop and the W-Φ motif. The distal loop, however, remains incorrectly aligned, mostly because of the influence of 1BB9 that contains a long insertion. |