|
|
|
|
|
Expresso and 3D-Coffee
Combining Protein Sequences and Structures |
|
|
|
|
|
What are 3D-Coffee and Expresso?
|
|
|
|
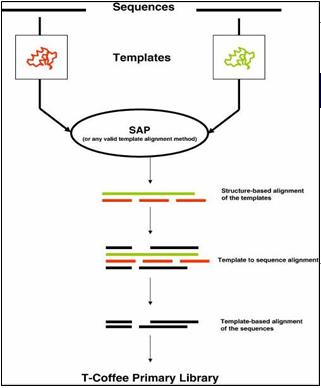
3D-Coffee is a special mode of T-Coffee that uses structural information. In 3D-Coffee the original sequences are replaced by homologous structures (templates). The templates are aligned using strutural aligners like sap or TMalign and these structure based alignments are used as a template for realigning the original sequences. Provided the right templates have been found, the result is a structure based multiple sequence alignment. If some of the sequences do not have any template, they are treated like regular sequences and aligned with the appropriate methods.
Expresso is an extension of 3D-Coffee where the structural templates are automatically identified with BLAST.
A list of related publications is available here
and the principle of the algorithm is explained in the following presentation.
|
|
|
|
|
|
|
|
|
|
- Expresso and 3D-Coffee are special modes of T-Coffee. Download the latest T-Coffee version here.
|
|
|
|
|
|
1-Follow the standard T-Coffee installation procedure.
|
|
2-Install all the structure alignment packages you want to combine. A list of these packages and their download address is available here.
You should also take a look at the T-Coffee Technical Documentation).
The latest list of packages interfaced with T-Coffee can also be obtained with the simple intruction t_coffee > package_list
|
|
|
|
|
|
|
|
When running on a foo.pep dataset, Expresso and 3D-Coffee output a file named foo.html that contains a color coded indication of the agreement between all the various pairwise structural alignments (i.e. an estimation of how consistent is the alignment of every sequence triplet AB, BC and AC) . Red brick regions are in perfect agreement across all the methods, while blue regions have a very poor agreement.
Red brick regions can safely be assumed to be correct while blue regions should be discarted. Green and Yellow regions should also be used with caution, especially when reconstructing a tree or building a profile. The top of the output provides the average conistency (0-100) for each sequence, thus indicating wether some sequences may have a less reliable alignment. A score lower than 50 should be considered poor.

|
|
EXPRESSO also outputs a file named foo.template_list that indicates which PDB template was associated to each original sequence. This file is a regular FASTA file, with the templates declared in the header (_P_ footemplate). You can modify this file and re-use it with the -template_file flag.

|
|
|
|
The full documentation is on the T-Coffee Homepage. But the following shortcuts may be useful.
|
| To run all the available methods, type |
t_coffee foo.seq -mode expresso
t_coffee foo.seq -mode 3dcoffee
|
|
| To use your own template file: |
t_coffee foo.seq -template_file foo.template_list -method sap_pair proba_pair
|
|
|
|
|
|
|
|
|
|
Our projects relie on your feeback. Please send me an
E-mail if you wish to make a request, a comment, or report a bug!
*******************************************
Dr. Cedric Notredame, PhD.
Group Leader
Comparative Bioinformatics Group
Bioinformatics and Genomics Programme
Center for Genomic Regulation (CRG)
Dr Aiguader, 88
08003 Barcelona
Spain
Email: cedric.notredame@gmail.com
HOME : http://www.tcoffee.org/
GROUP: CRG
Phone: +34 933 160 271
*******************************************
|
|
|
|