|
|
|
|
|
SARA-Coffee:
accurate multiple RNA structural alignment |
|
|
|
|
|
What is SARA-Coffee?
|
SARA-Coffee is a structure based multiple RNA aligner. This is a new algorithm that joins the pairwise RNA structure alignments performed by SARA with the multiple sequence T-Coffee framework.
SARA-Coffee is part of the T-Coffee distribution.
The SARA-Coffee paper.
Using tertiary structure for the computation of highly accurate multiple RNA alignments with the SARA-Coffee package.[medline][pdf]
|
|
|
From here it is possible to download theoriginal SARA-Coffee package (as used in the paper), a wrapper of perl and pyton scripts to reproduce the benchmak described in the SARA-Coffee paper.
- Download the SARA-Coffee benchmark package here
|
|
|
|
|
|
|
|
|
In order to run SARA-Coffee, the installation of both SARA and T-Coffee are needed.
SARA is availabe here, while
T-Coffee is available here
Alternatively, a pre-configured SARA-Coffee virtual machine is available for download at this link.
It allows you to test drive this method without having to install all the required packages manually.
|
|
|
|
|
|
|
|
THE STUCTURAL CONSISTENCY
|
SARA-Coffee stems from a combination SARA and T_Coffee packages, and is to our knowledge the first structure based multiple RNA aligner.
SARA performs a structural superposition using the C3’ carbons (on the ribose) as an alternative to the C-alpha protein superpositions are usually based upon. SARA then uses the resulting superposition to derive the equivalences needed to define a structure-based alignment. As such, SARA behaves like a standard aligner, thus making possible its incorporation in a consistency-based environment like T-Coffee.
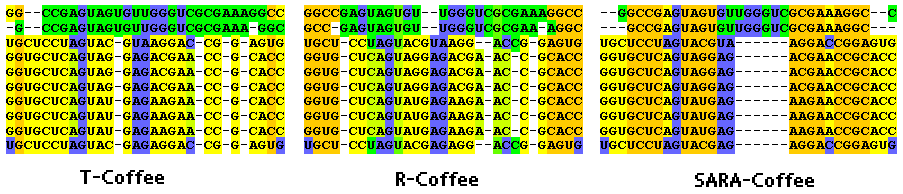
Multiple RNA alignments of the same sequence cluster. From left to right: T-Coffee, R-Coffee, SARA-Coffee. The color scheme reflects the 3SP score described in SARA-Coffee paper. This indicates the level of consistency between real secondary structures embedded in the alignment. Secondary structure in bad agreement are green, good ones orange. Blue colored nucleotides are not taking part in any base pairing.
|
|
|
|
To run SARA-Coffee are needed three components:
1) The RNA multiFASTA file to align
2) A folder containing all the considered RNA structures from PDB (Protein Data Bank)
3) A template file reporting the correspondence between the FASTA id and the PDB file name.
|
| To run SARA-Coffee type |
t_coffee -in <multiFASTA> -method saracoffee -template_file=TEMPLATEFILE
|
|
|
|
|
|
|
|
Our projects rely on your feeback. Please send an
E-mail if you wish to make a request, a comment, or report a bug!
*******************************************
Dr. Cedric Notredame, PhD.
Group Leader
Comparative Bioinformatics Group
Bioinformatics and Genomics Programme
Center for Genomic Regulation (CRG)
Dr Aiguader, 88
08003 Barcelona
Spain
Email: cedric.notredame@gmail.com
HOME : http://www.tcoffee.org/homepage.html
GROUP: CRG
Phone: +34 933 160 271
*******************************************
|
|
|
|
|