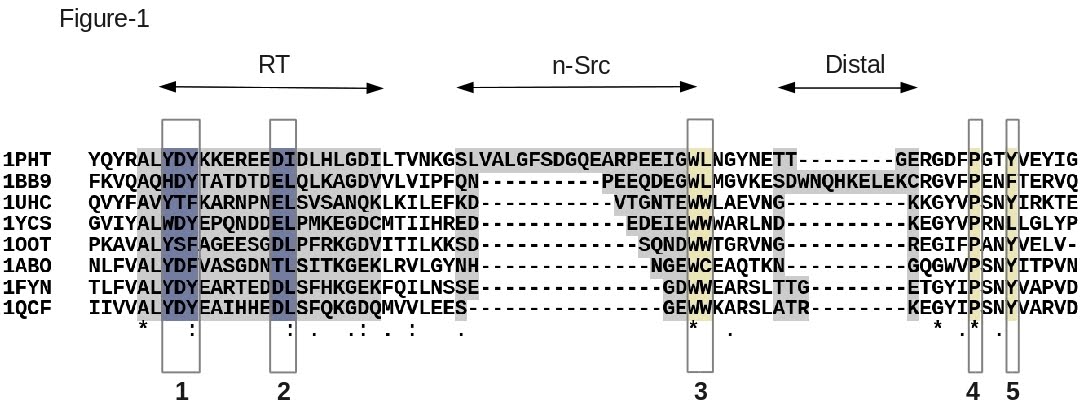
T-Coffee provides four different modes for protein alignments: T-Coffee (tcoffee default mode), Fast M-Coffee (fmcoffee), PSI-Coffee (psicoffee) and Expresso/3D-Coffee (expresso). Each of these modes uses different type of information: comparisons of sequences, pairwise sequence alignments, sequence profiles and protein structures. There is no 'ideal' mode and they all involve some trade-off between computational cost and alignment accuracy. The choice will therefore depend on the dataset size (Fast M-Coffee, T-Coffee), on the availability of structural information (Expresso, 3D-Coffee) and on the required level of accuracy (PSI-Coffee). In the following section we will be using these four flavors of T-Coffee for the alignment of a set of eight SH3 domains with known 3D structures. A reference alignment of these sequences has been established by specialists [32] (Figure 1) and will be used to estimate the accuracy of the output alignments.
|