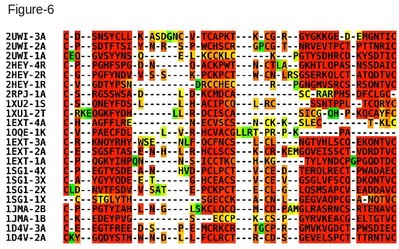
In this protocol we show how a structure based multiple sequence alignment of the Cystein Rich Domains (CRDs), from the tumor necrosis factor receptor family, can be computed with 3D-Coffee and used to do a structural classification of this domain family. This is a challenging dataset as it is made of very short domains with less than 20% identity. With such short and repeated domains, it is safer to manually identify the templates of each sequence. The following command line can therefore be used to generate the alignment of Figure 6. Note that in this example MUSTANG is used as pairwise method rather than SAP. t_coffee -seq crd.fasta -method mustang_pair -template_file crd.template_file Since every sequence in this alignment has a known 3D structure, structure based comparisons can be used to estimate the distance between every CRD pair. The T-RMSD method does this analysis by comparing intra-molecular distances between pairs of ungapped positions (in the MSA). In order to insure a meaningful clustering, it may therefore be useful to remove the sequences causing large number of gap insertions. Given any alignment containing sequences with a known 3D structure and the corresponding template file, T-RMSD runs with the following command: t_coffee -other_pg trmsd -aln crd.aln -template_file crd.template_file This command line produces four files:
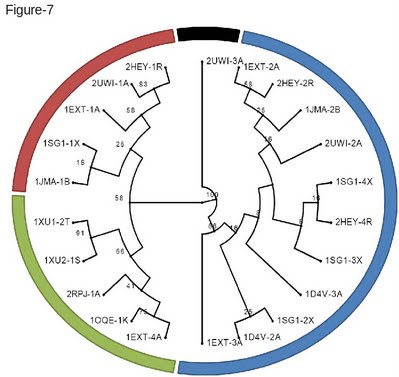
The Newick format is a standard tree format that can be visualized with online phylogenetic tools such as PhyloWidget [35] (Figure 7). On this clustering the numbers indicate the level of support for each corresponding node. This value is the equivalent of a bootstrap and corresponds to the fraction of ungapped columns effectively supporting a split. As shown on Figure 7, this structure based clustering is very consistent with the functional classification of these receptors and recapitulates most of the know functions associated with the CRD domain. |