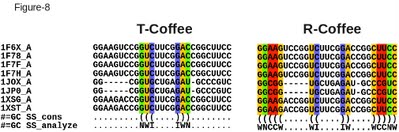
One of the most informative analysis when aligning RNA sequences is the identification of compensated mutations. Given an RNA alignment, with either a predicted or an estimated secondary structure, one can easily use the seq_reformat option of T-Coffee to estimate the number of columns showing compensated mutations. This is what the following command line will do on an alifold predicted secondary structure (the structure is predicted taking into account ungapped columns only). If available, an experimental (or alternative) structure may also be provided. t_coffee -other_pg=seq_reformat -in RNA_rcoffee.aln -action +alifold2analyze color_html > RNA_rcoffee.comp.html The resulting output is shown on Figure 8, both for R-Coffee and the default T-Coffee outputs. As one can see, the R-Coffee alignment contains four columns associated with compensated mutations. Such a finding is interesting as it yields increased support to the existence of a secondary structure shared by all the considered sequences. This finding also increases the level of confidence one may have in the considered alignment. This output can also be displayed in numerical form: t_coffee -other_pg=seq_reformat -in RNA_rcoffee.aln -action +alifold2analyze The result is a list summarizing the number of base pairs (bp) with compensated mutations. In the example presented here, 0 for T-Coffee and 2 in the R-Coffee alignment. |