T-Coffee usually provides a reasonable trade-off between speed and accuracy. It is significantly slower than the fastest available methods like Kalign, MAFFT or MUSCLE, but often more accurate on distantly related sequences. The accuracy and the CPU requirements of T-Coffee are comparable to those of ProbCons. However, T-Coffee runs in parallel mode and will therefore be significantly faster on a multi-core architecture [33]. Its usage is very straightforward: t_coffee -seq sh3.fasta T-Coffee produces three different files: - sh3.dnd: guide tree used to assemble the progressive alignment.
- sh3.aln: final alignment given in ClustalW format.
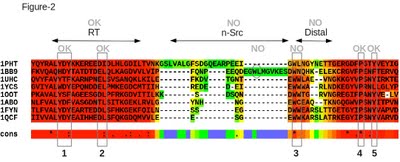
- sh3.html: final alignment colored in function of the CORE index from red (very consistent) to blue (poorly consistent) (Figure 2).
The alignment on Figure 2 has a score of 90, which means that it is 90% consistent with its primary library. The red bits in the alignment are expected to be the most accurate (according to the CORE index). A high consistency does not prove these portions to be correctly aligned, but it suggests they are more likely to be correctly aligned than the other positions with a lower score. Indeed, when comparing this MSA to the reference, one can see that the overall alignment seems to be correct with the exception of three functional regions: the W-Φ motif (Φ for hydrophobic), n-Src and distal loops. In particular, the motif W-Φ involved in the binding pocket (motif 3) is not correctly aligned as T-Coffee aligns the residues WN of the sequence 1BB9 instead of residues WL. Such a misalignment is hard to identify without structural information, especially when the inaccurate position contains a highly conserved tryptophan. |