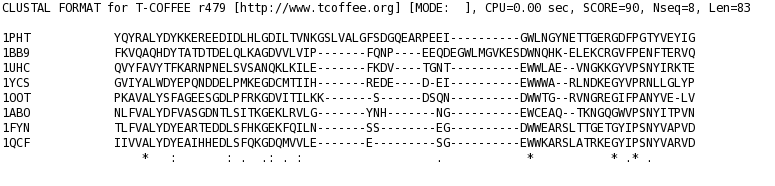
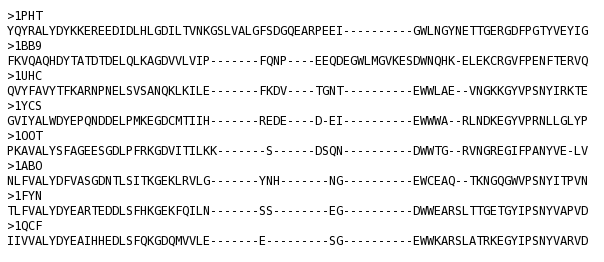
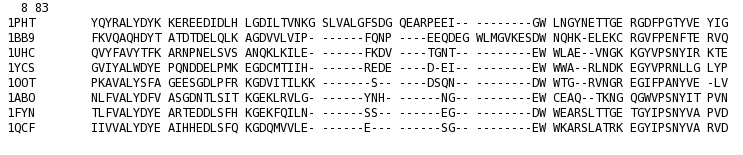
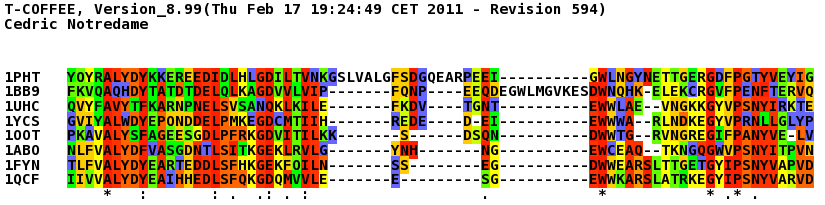
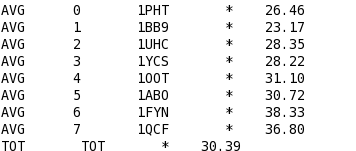
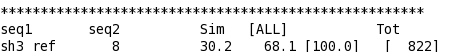
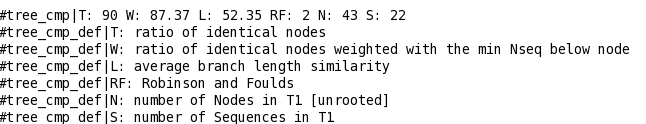
T_Coffee provides also a tool box for cleaning, trimming, reformating or comparing your data: fasta files, multiple sequences alignments, trees (Newick format), structure files etc... It can be used for pre-processing as post-processing steps. t_coffee -other_pg "command" 1) Changing the format of your sequences or alignments: "Changing from a clustalw format to fasta, phylip..." t_coffee -other_pg seq_reformat -in sh3.aln -output fasta_aln > sh3_fasta.aln t_coffee -other_pg seq_reformat -in sh3.aln -output phylip_aln > sh3_phylip.aln "Removing gaps from an alignment (MSA -> dataset) " t_coffee -other_pg seq_reformat -in sh3.aln -output fasta_seq 2) Trimming alignments: "Keep only sequences with less than 90% identity..." t_coffee -other_pg seq_reformat -in sh3.fasta -action +trim _seq_%%90_"Removing outliers with less than 30% identity with all sequences.." t_coffee -other_pg seq_reformat -in sh3.fasta -action +trim _seq_%%90_O30 "Keep only 10 sequences more informative..." t_coffee -other_pg seq_reformat -in sh3.fasta -action +trim _seq_n10_"Keep the 20% more informative sequences..." t_coffee -other_pg seq_reformat -in sh3.fasta -action +trim _seq_N20_ "Keep columns with less than 50% gaps..." t_coffee -other_pg seq_reformat -in sh3.fasta -action +rm_gap 50 3) Extracting information... "Extracting fasta sequence from a PDB file..." t_coffee -other_pg extract_from_pdb 1PHT.pdb -fasta "Extracting block 10 to 20 (respect consensus) ... " t_coffee -other_pg seq_reformat -in sh3.aln -action +extract_block cons 10 20"Extracting block 20 to 40 (respect seq 1PHT) ... " t_coffee -other_pg seq_reformat -in sh3.aln -action +extract_block '1PHT' 20 40 4) Coloring alignments t_coffee -other_pg seq_reformat -in sh3.aln -action +color_residue 1PHT 10 1 -output color_html > color.html"Coloring aligment from file ... " t_coffee -other_pg seq_reformat -in sh3.aln -action +color_residue color.txt -output color_html > file2color.html"Coloring by conservation ... " t_coffee -other_pg seq_reformat -in sh3.aln -in3 sh3.aln -action +3evaluate pam250mt -output color_html > cons2color.html "Coloring by boxshade... " t_coffee -other_pg seq_reformat -in sh3.aln -in3 sh3.aln -action +3evaluate boxshade -output color_html > boxshade2color.html 5) Comparing/Measuring quality, identity and similarity "Estimating diversity, average and pairwise identities between sequences in an alignment or dataset..." t_coffee -other_pg seq_reformat -in sh3.aln -output sim "Comparing alignments ... " t_coffee -other_pg aln_compare -al1 sh3_ref.aln -al2 sh3.aln -compare_mode column "Comparing trees ... " t_coffee -other_pg seq_reformat -in crd.struc_tree100 -in2 crd.struc_tree50 -action +tree_cmp |